[Originally published as Pollen Grain Size and Ploidy Level.
Editor’s note: While this is more technical than we usually publish, it serves several interesting purposes. First, this is real data collection that all scientists must do to discover what directions to take their future work, and it’s not difficult to skip to the end after learning about something as interesting to botanists as “ploidy” is.]
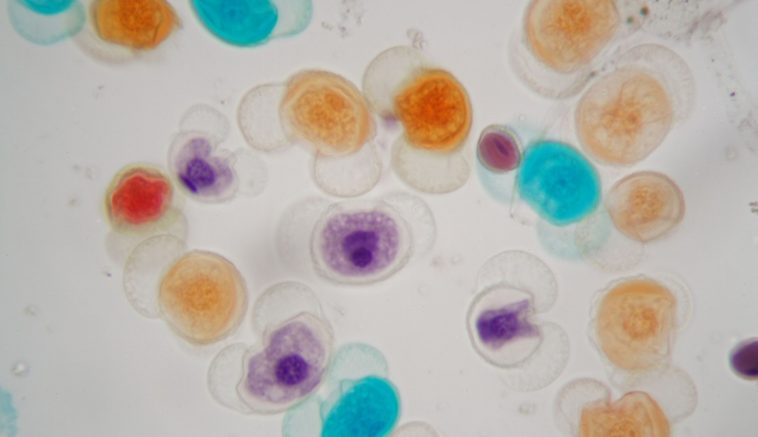
Pollen grain size has been regularly used as an indicator of the ploidy level in plants. It has been demonstrated in some plants and assumed in others that the larger the pollen, the higher the level of ploidy. However, very little work has been done to confirm that this rule holds in at least the majority of cases.
Most correlations of pollen grain size to ploidy level have been a side note in other research and have not considered the theoretical and genomic implications involved in such a measurement. While much of the literature presents ploidy level as being correlated with pollen grain size, there are some hints that the association might actually be with C-value. It is thus important to ensure that pollen grains are an accurate indicator of ploidy level if they are to continue to be used as such in the literature.
Introduction
Polyploidy, sometimes referred to as whole-genome duplication (WGD), is a common occurrence in plants. Exactly how common is debated, but conservative figures estimate that somewhere between twenty-four (Barker et al, 2016) and thirty-five percent (Wood et al, 2009) of extant plant taxa are recently formed polyploids. Polyploidy is largely clustered within angiosperm plants. Because a whole-genome duplication increases the amount of DNA in each cell, the size of the cells often increases to fit the additional DNA (Cavalier-Smith, 1978, Zhang et al, 2019). This increase in cell size is so well accepted that it has been used to estimate ploidy levels in fossil plants, by looking at the size of the fossil plant’s guard cells (Masterton, 1994).
Pollen grains are two-celled gametes released by angiosperms to fertilize either themselves or other members of the species. The gamete contains DNA, a single copy of the parent plant’s genome. This state, usually referred to as haploid and signified by n, is affected by polyploidy.
In even-numbered polyploids (tetraploids, octoploids, etc), the gametes will usually contain equal numbers of copies of the parental plant’s DNA, while in odd-numbered polyploids (triploids, etc) approximately half the gametes will have an odd number of copies of the DNA (the haploid n), while half will have an even number of copies (diploid 2n).
Such an increase in DNA size should cause an increase in the size of the pollen grains as well. Therefore, theoretically, polyploidy should increase the size of at least half of the pollen grains in any plant species and almost all pollen grains in even-numbered polyploids. This argument is common sense and is sometimes assumed in the literature (see Chung et al, 2013), but will not be assumed for the purposes of this review. Instead, this review attempts to determine if there is a correlation between pollen grain size and ploidy level.
Triploid Plants
Triploid plants, like other plants with odd numbers of chromosome copies, produce pollen during normal meiosis slightly differently than diploid or higher even-numbered ploidy level plants. Instead of producing pollen that is solely n or 2n, odd levels of ploidy produce gametes in equal measure. In other words, triploids produce some gametes that are normal haploid gametes and some that are the potentially enlarged diploid gametes. Odd-numbered ploidy levels higher than three would have the same effect.
As an example, Martin et al (2019) found that diploid plants of genus Annona produced pollen smaller than their triploid cousins on average. However, the margin of error on both the diploids and the triploids was large enough that both averages fell within the margin of error of the other average.
This result would seem to argue that changes to pollen grain sizes are random and not correlated to ploidy level. However, Martin et al did not distinguish between triploid pollen that was 2n and n. Including the n pollen with the 2n meant that the average was likely lower than the 2n pollen alone would have been. Thus, this study cannot be taken as strong evidence that ploidy level does not correlate with pollen grain size.
A study on Parnassia palustris appears to establish triploid pollen as the bridge between tetraploid and diploid pollen grains. Funamoto et al (2006) found that diploid pollen on average was the smallest, followed by triploid, then tetraploid pollen. Both tetraploids and diploids were within the margin of error of the triploid pollen, but not each other. Like Martin et al, Funamoto et al did not differentiate between 2n and n pollen grains from the triploids. The results thus landing squarely between the tetraploid and its 2n pollen and the diploid and its n pollen are hardly surprising.
Artificially Produced Polyploids
Inducing polyploidy by the application of the chemical colchicine is a well-established technique used frequently in the literature. Its products have been split out here because Ahmad et al (2018) points out that pollen grain size increases with the application of colchicine in Gladiolus grandiflorus. Since they did not correlate this increase with an increase in ploidy levels, it is safer to assume there is no correlation, and thus colchicine-induced polyploids will be treated separately from naturally occurring ones.
Omidbaigi et al (2010) examined the common spice basil (Ocimum basilicum L.). They applied colchicine to some basil plants and compared cell sizes, including pollen, from the resulting tetraploids as well as the original diploids. Tetraploid pollen was much larger than diploid pollen on average and the margins of error do not overlap. This was also true in Tragopogon. Tate et al (2009) showed that the mean pollen grain size was largest for colchicine-induced tetraploids, followed by natural tetraploids. Diploids were the smallest. Standard errors were very small and did not cause overlap between the groups. This paper seems to confirm that colchicine treatments cause greater cell enlargement than occurs naturally in polyploids.
Natural Tetraploids and Higher Ploidy Levels
In naturally produced tetraploids and other even ploidy levels, there is a range of variation in pollen grain size and overlap between ploidy levels. In Eriotheca species, pollen grain size differed strongly between diploid, tetraploid, and hexaploid individuals (Oliveira et al, 2013). In Rosa hybrida, 2n pollen was roughly 30% larger on average than n pollen (Zhou et al,
2019). Likewise, while the tetraploid pollen was not double the size of the diploid pollen, it was larger in the Bouteloua curtipendula complex (Siqueiros-Delgado et al, 2017). These lines of evidence would seem to indicate that pollen size does increase with increasing ploidy levels.
Increasing the ploidy level does not always correlate exactly with increased pollen grain size. Tetraploid pollen was also larger than diploid pollen in Eragrostis tef, with no overlap between the two. (Gungsa and Loerz, 2013). However, octoploid pollen overlapped with tetraploid pollen while also presenting larger pollen grains. This pattern also held in Physochlaina praealta (Singhal et al, 2017a).
Tetraploid pollen was produced in three sizes, with the largest grains being much larger than the diploid pollen and outside the margin of error. However, the typical size overlapped, and smaller tetraploid pollen was much smaller than the diploid pollen average and outside the margin of error. The overlapping pattern in P. praealta was also seen in Inula grandiflora, except there was no “small” category in that species (Singhal et al 2017b).
The study in Physochlaina praealta, in particular, is confusing. Tetraploid plants producing pollen the same size, larger, and smaller than the diploid plant should not be the case if increasing levels of ploidy also increase pollen size. Such a result could be seen as refuting any correlation between pollen grain size and ploidy level, at least as an across-the-board rule. However, a possible solution suggests itself. The sizes of the pollen might be representative of the ploidy level. In other words, the largest pollen might be 2n, the typical size might be n, and the smallest pollen 0n due to errors in meiosis. If that were the case, however, pollen fertility would be expected to drop. It did not do so in tetraploids in Physochlaina praealta (Singhal et al, 2017a). Therefore, this explanation seems unlikely in this instance.
Objections to Pollen Size Increase
While pollen grain size has been frequently used to determine ploidy levels in the literature, there have been a few questions raised as to its value. Knight et al (2010) attempted to correlate pollen size to ploidy level with the hope to determine ploidy levels in fossils and was unable to do so. Knight et al’s analysis was heavily dependent on C-values (a measure of the mass of genome content) to determine the ploidy level.
This creates two problems. The first is the possibility that C-value does not correlate with the ploidy level. Given that species with the same ploidy level in the same genus have widely different C-values (see examples in Suda et al 2006), the assumption that a higher C-value is equivalent to a higher ploidy level cannot be substantiated, at least across all polyploid plants. The second problem comes from the tendency of polyploids to undergo gene loss over time (Buggs et al, 2009). Thus, while C-value might be a fair assessment of the ploidy level when the polyploid lineage is first formed, it becomes a much worse assessment over time, something Knight et al’s analysis did not take into account.
Another study looked at ploidy levels, C-value, and pollen grain size in Plantago (Wong and Murray, 2012). Like Knight et al, they found that there was no direct correlation between pollen size and ploidy level. However, they found that C-value correlated directly with pollen size. Given the gene loss polyploids undergo over time and the fact that Plantago is believed to be an older polyploid, this conclusion seems reasonable. It does, however, open the question of how reliable pollen size is as an indicator.
Other studies have noticed that pollen size is not always correlated to the ploidy level. Horandl et al (2017) discovered that pollen size in Ranunculus kuepferi was not an indicator of the ploidy level. In fact, tetraploids produced the majority of the smallest pollen in the study, something that clearly seems to go against what has been found elsewhere. A similar pattern occurred in Ramonda, where, despite differing ploidy levels, there was no distinct difference in pollen grain size (Siljack-Yakovlev et al, 2008). While in line with Horandl et al’s results, these results do not mesh with the literature expectations.
Conclusions
The above-presented data may seem very contradictory, and at first glance, it is. Some plants produce pollen that correlates nicely with ploidy level. Others produce pollen that does not correlate with the ploidy level. In many cases, there is an overlap between the sizes of the pollen at various ploidy levels. There are several possible explanations for this conflicting mass of data.
The first explanation is that there is a correlation between pollen grain size and ploidy level and that the counterexamples presented above are aberrations or incorrect. This argument would be in line with the accepted understanding in the literature, along with the observation that cell size increases leaf size in Populus (Zhang et al, 2018). However, it fails to provide a comprehensive explanation for variations in pollen grain size. Essentially, this explanation is content with explaining most, but not all, of the data. As such, other options should be considered.
It is possible that the reason some higher ploidy level plants do not have enlarged pollen is due to a selection-imposed limit to growth. In other words, selection might impose a constraint on the increase in pollen size. Tate and Simpson (2004) propose this idea for plants of genus Tarsa. They argue that selection has caused tetraploid pollen to shrink in average size over time to aid in reproduction. Such a selection constraint might be in place in some of the other aforementioned examples, explaining why pollen grains do not always correlate with ploidy levels, such as in Gungsa and Loerz’s (2013) work with Eragrostis.
Given that it has been proposed that self-compatibility can help jumpstart many polyploid species once they form (Horandl, 2008), it makes sense that pollen grains of polyploids might be under selective pressure to still fit in the stigma of their parent plants. Being able to reproduce is key to the fitness of an organism, so selection should favor pollen grain sizes that will permit the plant to produce offspring. However, as these structures are often enlarged, selection should not strongly impact pollen grain size in a self-compatibility scenario.
The issue with the selection explanation is that self-compatibility has been shown to not be directly correlated with polyploidy (Mable, 2004). Therefore, when a new polyploid arises, it may be self-incompatible. If it is self-incompatible, selection should immediately favor smaller pollen grains to fit its diploid relatives. Yet, in general, ploidy does seem to be very loosely correlated with pollen grain size. This would hint at least that selection may not be the driving force across the board in pollen grain size determination.
A third possible explanation is that pollen grain size does not correlate with the ploidy level at all. Instead, pollen grain size is correlated with C-value. Wong and Murray (2012) provide good evidence to support this argument. However, Dewitte et al (2009), specifically state that some members of genus Begonia have much different pollen grain sizes than other members of the genus with the same genome size. If pollen grain size correlated with C-value, a measure of genome size, then members of the same genus with the same genome size should have very similar pollen.
Another possible explanation is that there is no single universal factor undergirding pollen grain size. Instead, pollen grains size is controlled by a combination of selection constraints and C-value or ploidy level. This explanation seems perhaps the most plausible, but little work has been done to test it. Before concluding it makes sense, empirical testing must be done to ensure it has explanatory power.
There is much work to be done in this area. As seen in this review, pollen grain size does not seem to be explained by any single overriding factor. This being the case, it would be wise for researchers to refrain from using it as an indicator for the ploidy level until a correlation between the two can be established. If it must be used, it would be wise to check the literature to ensure that the ploidy level does correlate to pollen grain size in closely related species. However, even this is not a guarantee as the aforementioned Begonia study pointed out (Dewitte et al, 2009). Ideally, other markers should be used to determine ploidy level, as pollen grain size does not appear to be as reliable as might be expected.
References:
Ahmad T, Manzoor A, Bashir MA, Baig MMQ, Quresh AA, Shah MKN, Hafiz IA. 2018.
Induction and identification of colchicine induced polyploidy in Gladiolus grandifloras ‘white prosperity’. Folia Hort. 30(2):307-319.
Barker MS, Arrigo N, Baniaga AE, Li Z, Levin DA. 2015. On the relative abundance of autopolyploids and allopolyploids. New Phytol. 210(2):391-398.
Buggs RJA, Doust AN, Tate JA, Koh J, Soltis K, Feltus FA, Paterson AH, Soltis PS, Soltis DE. 2009. Gene loss and silencing in Tragopogon miscellus (Asteraceae): comparison of natural and synthetic allotetraploids. Heredity. 103:73-81.
Cavalier-Smith T. 1978. Nuclear volume control by nucleoskeletal DNA, selection for cell volume and cell growth rate, and the solution of the DNA C-value paradox. J Cell Sci. 34:247278.
Chung MY, Chung JD, Ramana M, van Tuyl JP, Lim KB. 2013. Production of polyploids and unreduced gametes in Lilium auratum x L. henryi hybrid. Int J Biol Sci. 9(7):693-701.
Dewitte, A, Leus L, Eeckhaut T, Vanstechelman I, Van Huylenbroeck J, Van Bockstaele E.
2009. Genome size variation in Begonia. Genome. 52(10):829-838.
Funamoto T, Kondo, K, Tatarenko IV, Gontcharov A, Verkholat VP, Smirnov SV. 2006. Intraspecific polyploidy of Parnassia palistris var multiseta (Saxifragaceae s. l.) collected in
Primorye and Altai Territories, Russia. Chromosom Bot. 1(1):23-26.
Gungsa L, Loerz H. 2013. Male sterility and gametoclonal variations from gynogenically derived polyploids of tef (Eragrostis tef), zucc Trotter. Afr J Plant Sci. 7(2):53-60.
Horandl E. 2008. Evolutionary implications of self-compatibility and reproductive fitness in the apomictic Ranunculus auricomus polyploid complex (Ranunculaceae). Int J Plant Sci.
169(9):1219-1228.
Horandl E, Schinkel CCF, Kirchheimer B, Dullinger S, Geelen D, De Storme N. 2017. Pathways to polyploidy: indications of a female triploid bridge in the alpine species Ranunculus kuepferi
(Ranunculadceae). Plant Syst Evol. 303:1093-1108.
Knight CA, Clancy RB, Gotzenberger L, Dann L, Beaulieu JM. 2010. On the relationship between pollen size and genome size. J Bot. 1-7.
Mable BK. 2004. Polyploidy and self-compatibility: is there an association?. New Phytol.
162(3):803-811.
Martin C, Viruel MA, Lora J, Hormaza JI. 2019. Polyplody in fruit tree crops of the genus Annona (Annonaceae). Front Plant Sci. 11.
Masterton, J. 1994. Stomatal size in fossil plants: evidence for polyploidy in majority of angiosperms. Science. 264(5157):421-424.
Oliveira PE, Marinho RC, Mendes-Rodrigues C, Bonetti AM. 2013. Pollen and stomata morphometrics and polyploidy in Eriotheca (Malvaceae-Bombacoideae). Plant Biol (Stuttg).
16(2):508-511.
Omidbaigi R, Mirzaee M, Hassani ME, Moghadam MS. 2010. Induction and identification of polyplody in basil (Ocimum baslicum L.) medicinal plant by colchicine treatment. Int J Plant Prod. 4(2):89-98.
Tate JA, Simpson BB. 2004. Breeding system evolution in Tarsa (Malvaceae) and selection for reduced pollen grain size in the polyploid species. Am J Bot. 91(2):207-213.
Tate JA, Symonds VV, Doust AN, Buggs RJA, Mavrodiev E, Majure LC, Soltis PS, Soltis DE.
2009. Synthetic polyploids of Tragopogon miscellus and T. mirus (Asteraceae): 60 years after
Owenbey’s discovery. Am J Bot. 96(5):979-988.
Siljak-Yakovlev S, Stevanovic V, Tomasevic M, Brown SC, Stevanovic B. Genome size variation and polyploidy in the resurrection plant genus Ramonda: cytogeography of living fossils. Environ Exp Bot. 62(2):101-112.
Singhal VK, Tantray YR, Kaur D, Gupta RC. 2017a. First report of intraspecific polyploidy (2x, 4x) in Physochlania praealta (Decne.) Miers. (family: Solanaceae). Cytologia. 82(3): 245-250.
Singhal VK, Himshikha, Gupta RC, Kumar R. 2017b Cytomixis and intraspecific polyploidy
(2x, 4x) in Inula grandiflora Willd. from Malana Valley, Kullu District, Himachal Pradesh.
Cytologia. 82(3):273-278.
Siqueiros-Delgago ME, Fisher AE, Columbus JT. 2017. Polyploidy as a factor in the evolution of the Bouteloua curtipendula complex (Poaceae: Chloridoideae). Syst Bot. 43(3):432-448. Suda J, Krahulcova A, Travnicek P, Krahulec F. 2006. Ploidy level versus DNA ploidy level: an appeal for consistent terminology. Taxon. 55(2):447-450.
Wong C, Murray B. 2012. Variable changes in genome size associated with different polyploid events in Plantago (Plantaginaceae). J. Hered. 103(5):711-719.
Wood TE, Takebayashi N, Barker MS, Mayrose I, Greenspoon PB, Rieseberg L. The frequency of polyploid speciation in vascular plants. Proc Natl Acad Sci U S A. 106(33):13875-13879. Zhang J, Zhang Y, Wang B, Qi S, Dong M, Wang Z, Li Y, Chen S, Li B. 2019. Ploidy and hybridity effects on leaf size, cell size, and related genes expression in triploids and their parents in Populus. Planta. 249:635-646.
Zhou Y, Gao S, Yang M, Zhang F, Fan L. 2019. The strong competitive role of 2n pollen in several polyploidy hybridizations in Rose hybrida. BMC Plant Biol. 19:127.