In his book, Why Evolution is True, biologist Jerry A. Coyne describes modern evolutionary theory with the following statement:
“Life on earth evolved gradually beginning with one primitive species—perhaps a self-replicating molecule—that lived more than 3.5 billion years ago; it then branched out over time, throwing off many new and diverse species.”
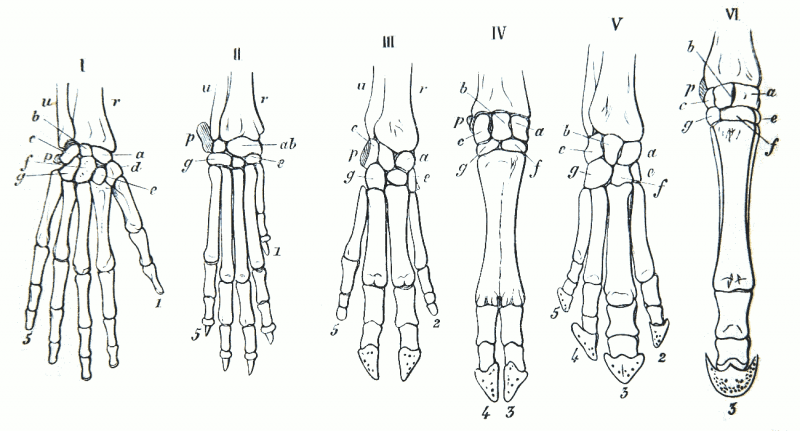
Based on this foundational principle, various species are expected to share similar (or homologous) structures due to common ancestry. Berkeley’s Evolution 101 page notes, “Evolutionary theory predicts that related organisms will share similarities that are derived from common ancestors. Similar characteristics due to relatedness are known as homologies.”
Wikipedia’s homology entry lists the following example: “…the forelimbs of vertebrates, where the wings of bats, the arms of primates, the front flippers of whales and the forelegs of dogs and horses are all derived from the same ancestral tetrapod structure.”
However, the relatively recent advent of DNA sequencing has produced some very unexpected results- surprisingly contrary to evolutionary theory predictions.
DNA Sequencing- We’ve Come a Long Way Baby
DNA is the blueprint, or instruction manual, containing the instructions which make every species unique. DNA (pictured below) is defined as, “…a thread-like chain of nucleotides carrying the genetic instructions used in the growth, development, functioning and reproduction of all known living organisms and many viruses.”
In the 1960’s scientists developed the ability to “read” this DNA instruction manual in a process called DNA sequencing. This was a monumental scientific breakthrough. Britannica defines DNA sequencing and notes its significance with the following entry:
“…technique used to determine the nucleotide sequence of DNA… The nucleotide sequence is the most fundamental level of knowledge of a gene or genome. It is the blueprint that contains the instructions for building an organism, and no understanding of genetic function or evolution could be complete without obtaining this information.” (emphasis mine)
The last sentence is imperative. While Darwin and his predecessors could hypothesize that common ancestry is the most reasonable explanation for similarities shared among various species based on visual comparison, common ancestry cannot be proven and the very concept of evolution cannot be understood without the ability to “read” an organism’s instruction manual (DNA). This capability is very new to science.
For several decades DNA sequencing was a slow and expensive process. However, the Human Genome Project, initiated in 1990 and completed in 2003, had a revolutionary effect. The goal of this international project, to map the entire human genome, spurred tremendous technological advances in gene sequencing which has continued far beyond the project’s completion. James Heather concludes his History of Sequencing DNA by stating, “Over the years, innovations in sequencing protocols, molecular biology and automation increased the technological capabilities of sequencing while decreasing the cost, allowing the reading of DNA hundreds of basepairs in length, massively parallelized to produce gigabases of data in one run.”
It is this burgeoning wealth of genetics information that has revealed the “mystery” of orphan genes.
What are Orphan Genes and Why are They Problematic for Evolutionary Theory?
Cornelius Hunter, writing for Evolution News, provides the following definition, “The term orphan refers to a DNA open reading frame, or ORF, without any known similar sequence in other species or lineages. Hence ORFan, or ‘orphan.’” The author of this article in Uncommon Descent supplies this definition, “Orphan genes are presumed protein coding genes that exist in only one species and have such non-similarity to anything in any other species they are called orphans…”
Why is this troubling? If the theory of evolution and (by default) common ancestry are true, a coding gene that is species specific, with no recognizable counterpart in other species should be an extreme rarity. Ann Gauger writes in Orphan Genes: A Guide for the Perplexed, “The working assumption had been that, given common descent and the fact that most housekeeping genes are shared among living things, and the assumption hitherto that evolution occurs by incremental small changes, orphan genes…should be rare if not non-existent.”
So, just how common are they? This 2009 study published in Trends in Genetics found, “Comparative genome analyses indicate that every taxonomic group so far studied contains 10-20% of genes that lack recognizable homologs in other species.” According to Richard Buggs (writing for Ecology and Nature), researchers originally believed that the mystery of these orphan genes would be resolved over time as more genomes were sequenced, finding precursors for the sequences that are now categorized as orphans. However, the opposite has proven true.
For example, Dr. Jeffrey Tompkins discusses ants, “When comparing the ant genes to other insects, researchers discovered 28,581 genes that were unique only to ants and not found in other insects. While the various ant species shared many groups of genes, only 64 genes were common to all seven ant species…The researchers concluded that on average, each ant species contained 1,715 unique genes—orphan genes.”

In Buggs’ 2017 ash tree genome paper published in Nature, he and his colleagues report that of the over 38,000 protein-coding genes found, “…one quarter (9,604) were unique to ash.” He continues, “On the basis of our research so far, I cannot suggest shared evolutionary ancestry for these genes with those in ten other plants we compared ash to: coffee, grape, loblolly pine, monkey flower, poplar, tomato, Amborella, Arabidopsis, barrel medic, and bladderwort. This is despite the fact that monkey flower and bladderwort are in the same taxonomic order (Lamiales) as ash.”
Not only are orphan genes common, they also appear to be functional. Dr.Tompkins writes, “These orphan genes are also being found to be particularly important for specific biological adaptations that correspond with ecological niches in relation to the creature’s interaction with its environment. The problem for the evolutionary model of animal origins is the fact that these DNA sequences appear suddenly and fully functional without any trace of evolutionary ancestry (DNA sequence precursors in other seemingly related organisms).”
Conclusion
Orphan genes are certainly a fly in the ointment for evolutionary theory, but no surprise to either creation science or intelligent design. As Gauger points out, “Then there is the elephant in the room that evolutionary biologists don’t want to acknowledge. Perhaps we see so many species- and clade-specific orphan genes because they are uniquely designed for species- and clade-specific functions. Certainly, this runs contrary to the expectation of common descent.”
In Part 2 of this series, we’ll take a look at how evolutionary biology responds to orphan genes.







